Structural Variants¶
Structural variants should be passed to the IPR python adapter in the main report content JSON.
{
"structuralVariants": [
// variants
]
}
Each variant is an object which may contain any of the following fields
| Field | Type | Example | Description |
|---|---|---|---|
| breakpoint (required) | string |
"12:123456|14:1244662" |
description of the breakpoints involved in this structural variant |
| eventType (required) | string |
"deletion" |
the type of underlying structural variant |
| exon1 (required) | integer? |
1 |
the 5' (n-terminal) exon |
| exon2 (required) | integer? |
2 |
the 3' (c-terminal) exon |
| gene1 (required) | string |
"EWSR1" |
the 5' (n-terminal) gene name |
| gene2 (required) | string |
"FLI1" |
the 3' (c-terminal) gene name |
| cTermTranscript | string? |
"ENST0004.5" |
the 5' transcript name |
| conventionalName | string |
cytogenetic descriptor | |
| detectedIn | string |
"DNA" |
the sample type(s) this SV was detected in |
| highQuality | boolean? |
This structural variant has a high level of supporting evidence | |
| nTermTranscript | string? |
"ENST0001.2" |
the 3' transcript name |
| omicSupport | boolean? |
flag to indicate this SV has supprt from both genome and transcriptome | |
| svg | string? |
svg image file content for this SV | |
| svgTitle | string? |
The title to accompany this SV |
Images¶
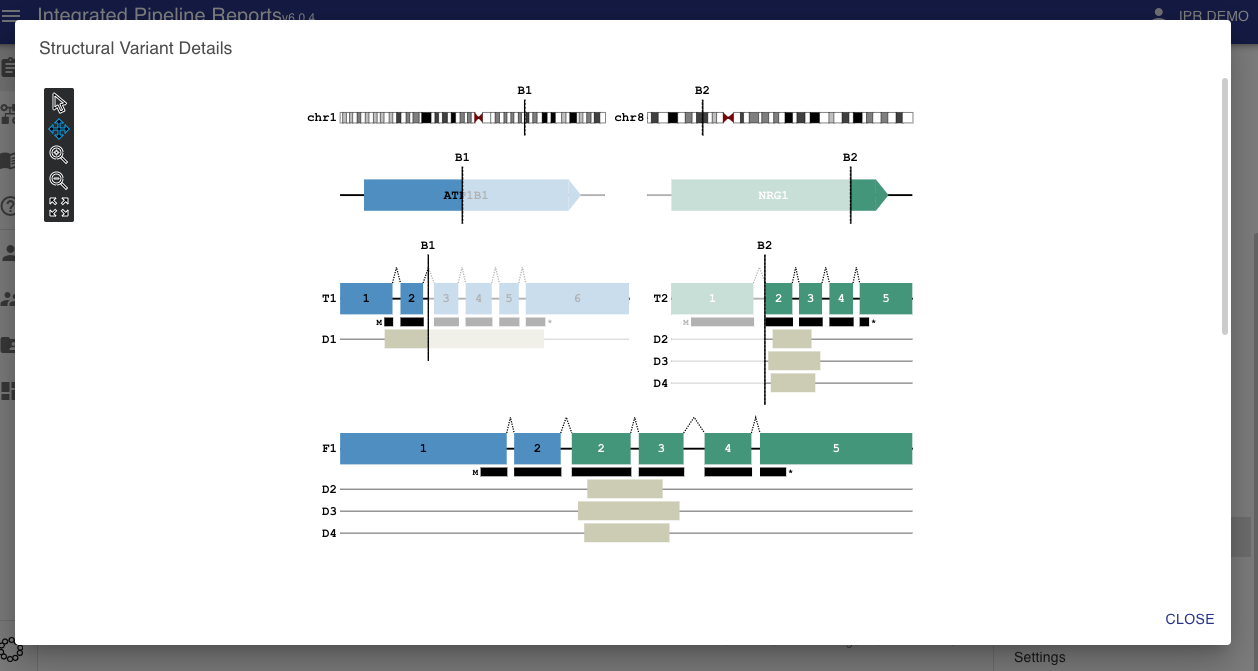
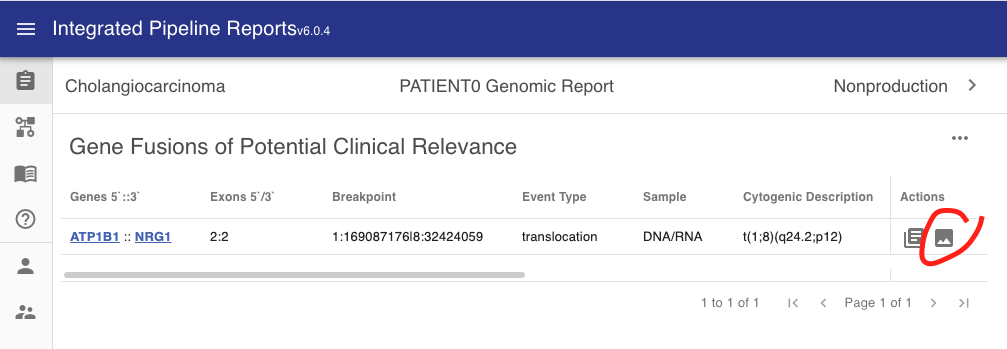
When the svg field contains an SVG image string this image will be displayed by the report via the actions tab (see button circled in red below). This allows the user to bring up the visualization when they are reviewing the structural variants in the report.
Clicking this button will bring the user to a pop up showing the visualization. The visualization shown below was created with MAVIS.