Gene Example¶
The get_equivalent_features() method is
used to find genes equivalent to the input/target feature.
from graphkb.match import get_equivalent_features
genes = get_equivalent_features(graphkb_conn, 'KRAS')
This will use a similar algorithm to what we have seen above in the disease matching example.
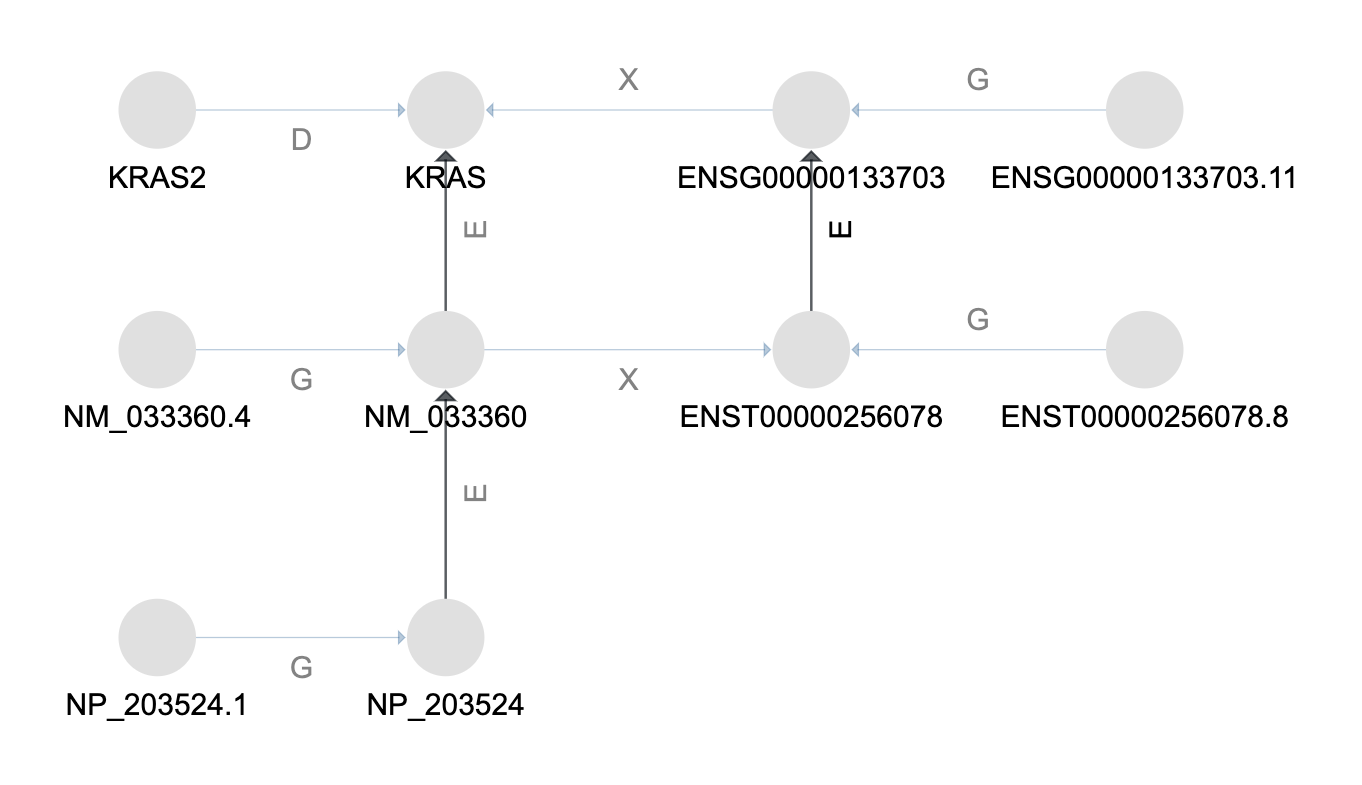
In the graph above the relationship types shown are: GeneralizationOf (G), ElementOf (E),
DeprecatedBy (D), and CrossReferenceOf (X).
Match by Name¶
As before, the first thing done is to match the input name
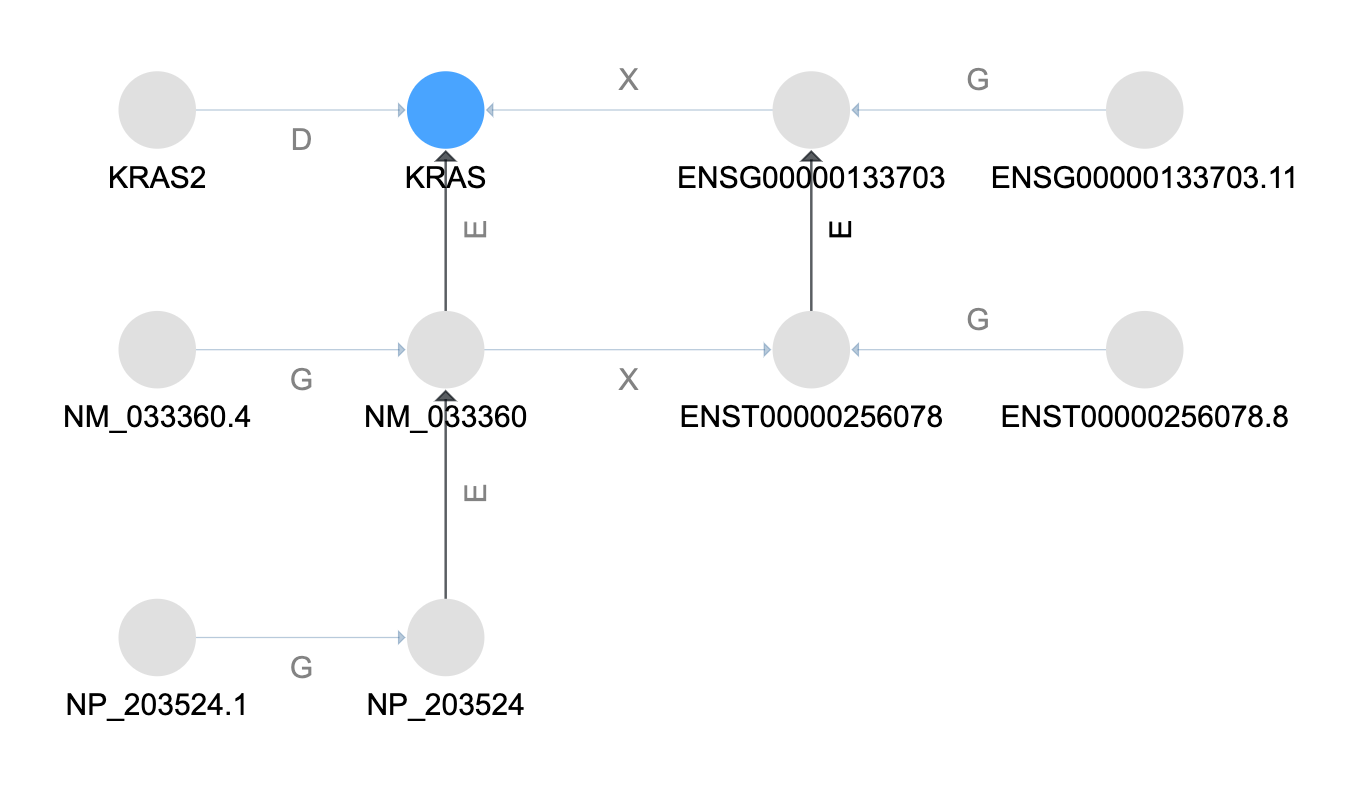
Resolve Aliases¶
The next step is to resolve equivalent names of the current set of terms.
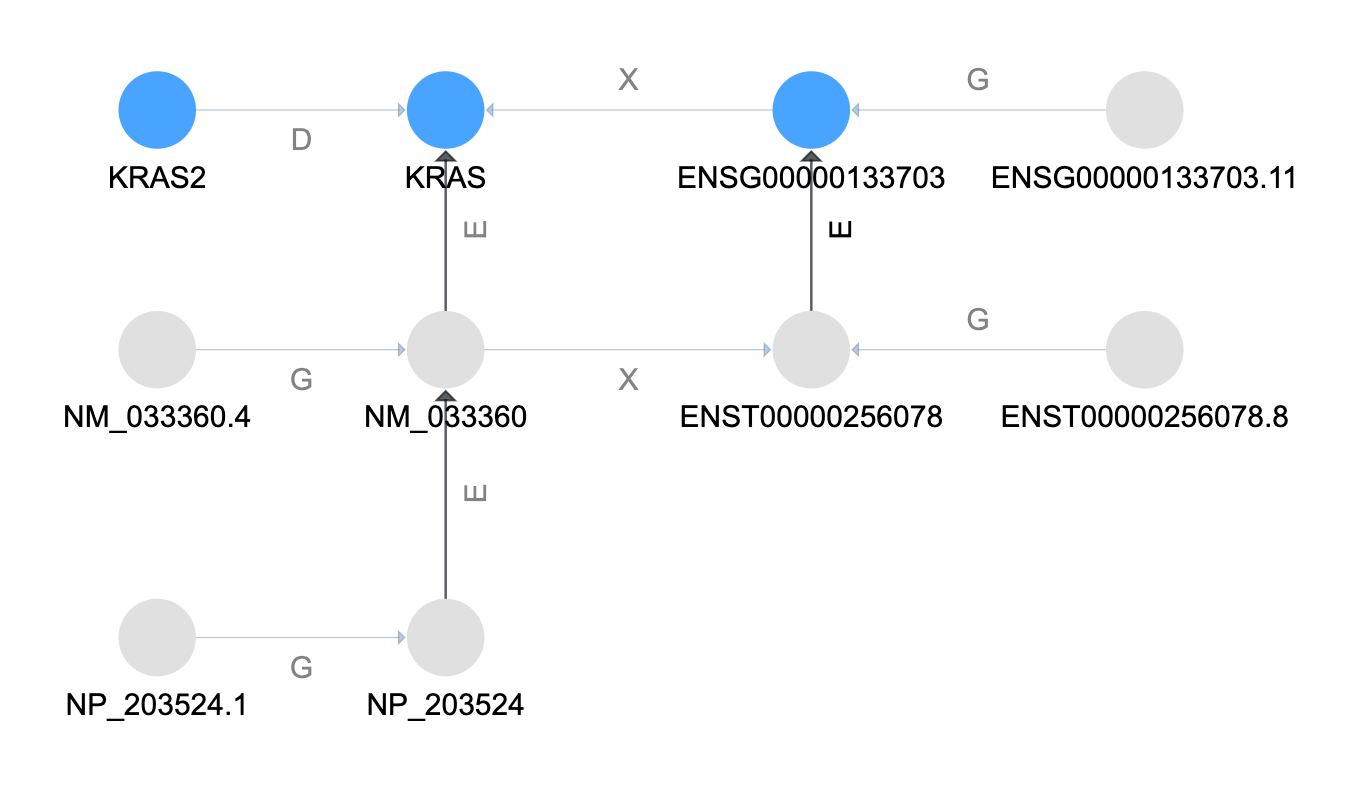
Follow the Elements Tree¶
The next step is to follow the element relationships. This is treated the same as the subclassing
except now our "tree edge" is the ElementOf relationship type.
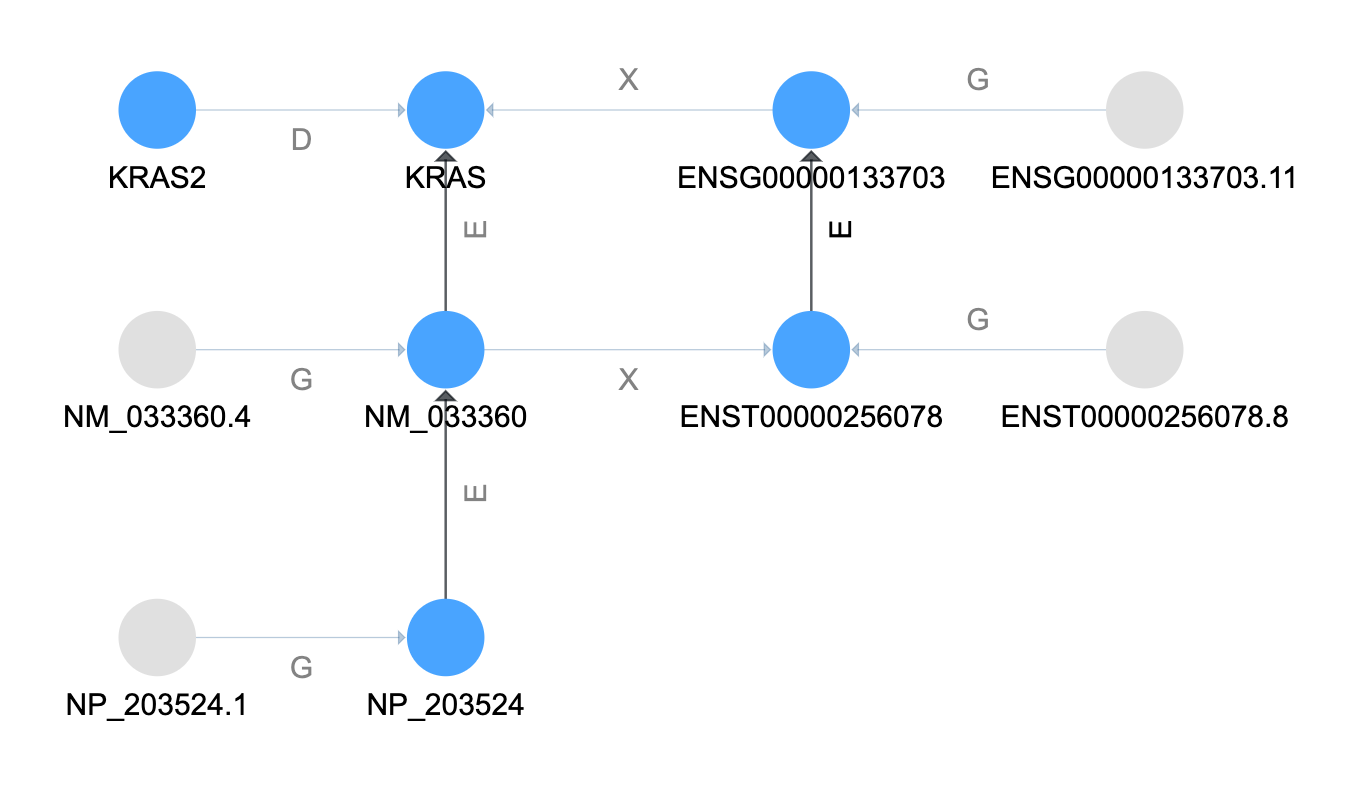
Resolve Final Aliases¶
Finally we expand the current set of terms by alias terms again to capture aliases of the more general parent and more specific child terms expanded in the previous step.
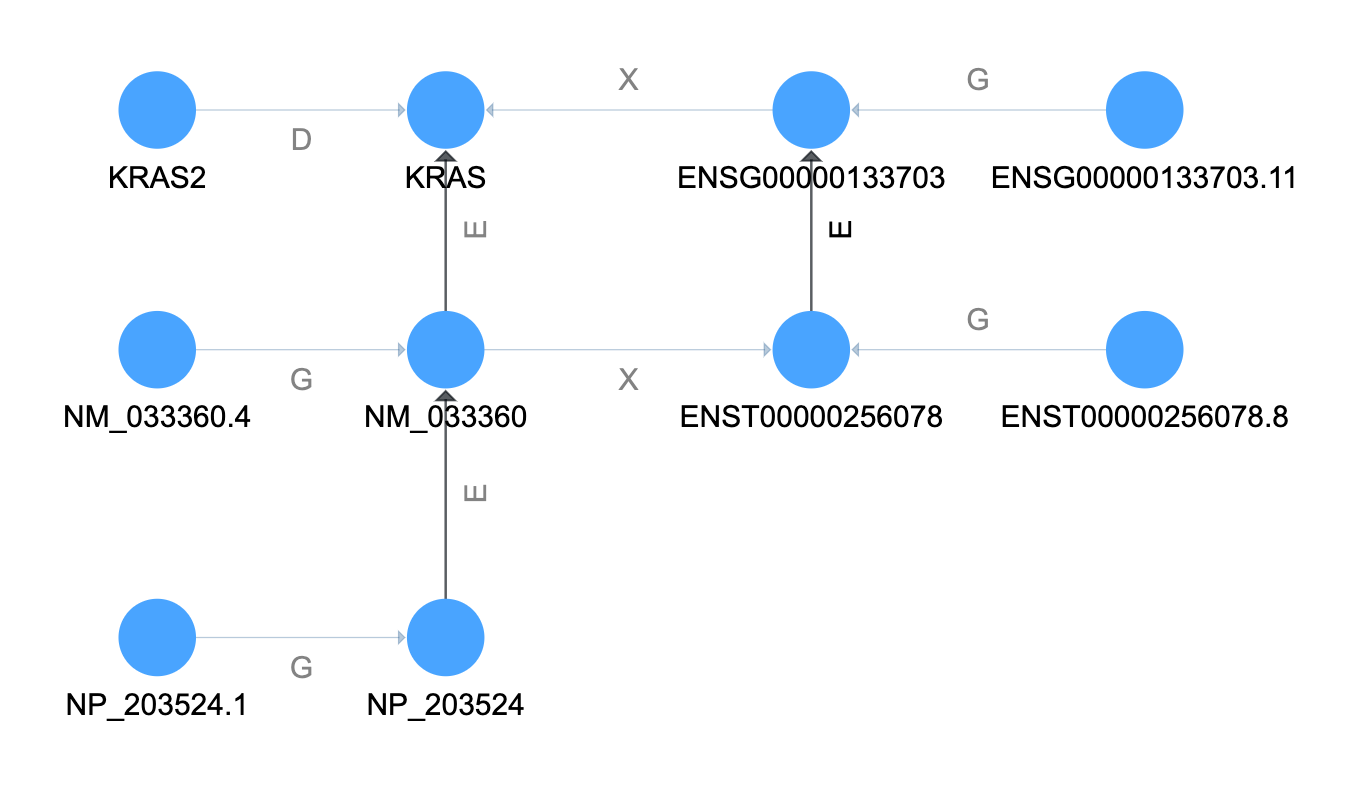
We have now collected all of the different terms for KRAS