Correct misassemblies in genome assembly drafts using linked or long sequencing reads
Cut sequences at positions with few spanning molecules.
Written by Shaun Jackman, Lauren Coombe, Justin Chu, and Janet Li.
Citation
Shaun D. Jackman, Lauren Coombe, Justin Chu, Rene L. Warren, Benjamin P. Vandervalk, Sarah Yeo, Zhuyi Xue, Hamid Mohamadi, Joerg Bohlmann, Steven J.M. Jones and Inanc Birol (2018). Tigmint: correcting assembly errors using linked reads from large molecules. BMC Bioinformatics, 19(1). doi:10.1186/s12859-018-2425-6
Description
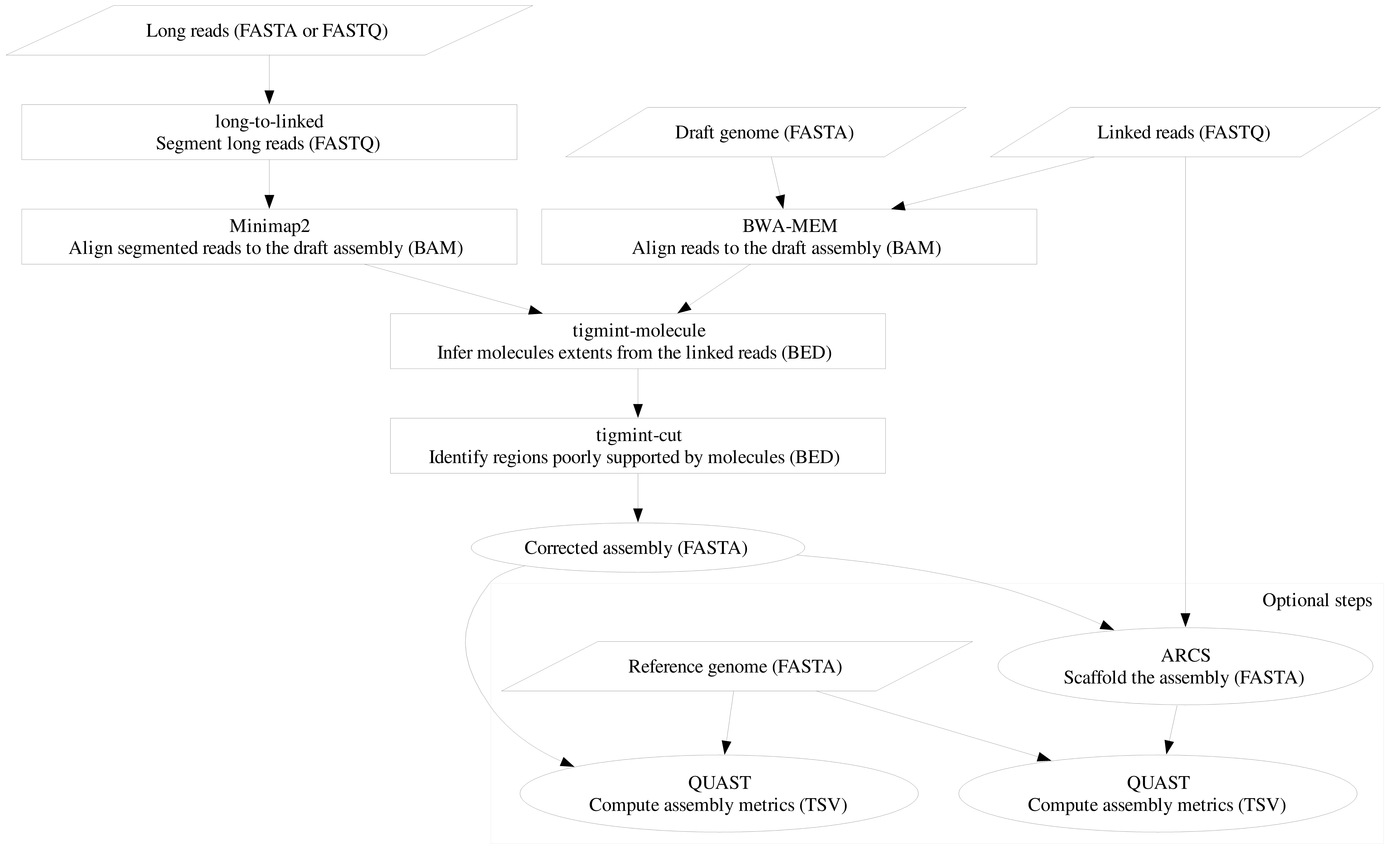
Tigmint identifies and corrects misassemblies using linked (e.g. MGI’s stLFR, 10x Genomics Chromium) or long (e.g. Oxford Nanopore Technologies long reads) DNA sequencing reads. The reads are first aligned to the assembly, and the extents of the large DNA molecules are inferred from the alignments of the reads. The physical coverage of the large molecules is more consistent and less prone to coverage dropouts than that of the short read sequencing data. The sequences are cut at positions that have insufficient spanning molecules. Tigmint outputs a BED file of these cut points, and a FASTA file of the cut sequences.
Tigmint also allows the use of long reads from Oxford Nanopore Technologies. The long reads are segmented and assigned barcodes, and the following steps of the pipeline are the same as described above.
Each window of a specified fixed size is checked for a minimum number of spanning molecules. Sequences are cut at those positions where a window with sufficient coverage is followed by some number of windows with insufficient coverage is then followed again by a window with sufficient coverage.
Installation
Install Tigmint using Brew
brew install tigmint
Install Tigmint using Conda
conda install -c bioconda -c conda-forge tigmint
Run Tigmint using Docker
docker run quay.io/biocontainers/tigmint
Install Tigmint from the source code
Download and extract the source code.
git clone https://github.com/bcgsc/tigmint && cd tigmint
make -C src
or
curl -L https://github.com/bcgsc/tigmint/releases/download/v1.2.10/tigmint-1.2.10.tar.gz | tar xz && cd tigmint-1.2.10
make -C src
Dependencies
Install Python package dependencies
conda install -c bioconda intervaltree pybedtools pysam numpy bedtools minimap2 bwa zsh btllib samtools
Install the dependencies of ARCS (optional)
conda install -c bioconda arcs links
Install the dependencies for calculating assembly metrics (optional)
conda install -c bioconda abyss seqtk
Usage
To run Tigmint on the draft assembly myassembly.fa with the reads myreads.fq.gz, which have been run through longranger basic:
tigmint-make tigmint draft=myassembly reads=myreads
bwa mem -Cis used to copy the BX tag from the FASTQ header to the SAM tags.samtools sort -tBXis used to sort first by barcode and then position.
To run both Tigmint and scaffold the corrected assembly with ARCS:
tigmint-make arcs draft=myassembly reads=myreads
To run Tigmint, ARCS, and calculate assembly metrics using the reference genome GRCh38.fa:
tigmint-make metrics draft=myassembly reads=myreads ref=GRCh38 G=3088269832
To run Tigmint with long reads in fasta or fastq format (myreads.fa.gz or myreads.fq.gz - or uncompressed) on the draft assembly myassembly.fa for an organism with a genome size of gsize:
tigmint-make tigmint-long draft=myassembly reads=myreads span=auto G=gsize dist=auto
minimap2 map-ontis used to align long reads from the Oxford Nanopore Technologies (ONT) platform, which is the default input for Tigmint. To use PacBio long reads specify the parameterlongmap=pb. The former callsminimap2 -x map-ontwhile the latter callsminimap2 -x map-pbinstead. When using PacBio HiFi long reads, specify the parameterlongmap=hifi.
Optionally, ntLink (v1.3.6+) can be used to map the long reads to the draft assembly. To use ntLink mappings, specify mapping=ntLink to your tigmint command.
Notes
tigmint-makeis a Makefile script, and so anymakeoptions may also be used withtigmint-make, such as-n(--dry-run).- When running Tigmint with linked reads, the file extension of the assembly must be
.faand the reads.fq.gz, and the extension should NOT be included in the parametersdraftandreadson the command line (otherwise you will get an error). These specific file name requirements result from implementing the pipeline in GNU Make.- The requirements for running Tigmint with long reads are the same, but the file extension of the reads file can also be
.fa,.fa.gz, or.fq - Ensure that the
draftandreadsnames are not identical
- The requirements for running Tigmint with long reads are the same, but the file extension of the reads file can also be
- The minimum spanning molecules parameter (
span) fortigmint-cutis heavily dependent on the sequence coverage of the linked or long reads provided. When running Tigmint with long reads, usespan=autoand setGto your assembly organism’s haploid genome size for this parameter to be calculated automatically, or explicitly setspanto a specific number if you are interested in adjusting it. See Tips for more details. - For
tigmint-long, the maximum distance between reads threshold should be calculated automatically based on the read length distribution. This can be done by setting the parameterdist=auto. - The
long-to-linked-pestep oftigmint-longuses a maximum of 6 threads
tigmint-make commands
tigmint: Run Tigmint, and produce a file named$draft.tigmint.fatigmint-long: Run Tigmint using long reads, and produce a file named$draft.cut$cut.tigmint.faarcs: Run Tigmint and ARCS, and produce a file name$draft.tigmint.arcs.fametrics: Run, Tigmint, ARCS, and calculate assembly metrics usingabyss-facandabyss-samtobreak, and produce TSV files.
Parameters of Tigmint
draft: Name of the draft assembly,myassembly.fareads: Name of the reads,myreads.fq.gzG: Haploid genome size of the draft assembly organism. Required to calculatespanparameter automatically. Can be given as an integer or in scientific notation (e.g. ‘3e9’ for human) [0]span=20: Number of spanning molecules threshold. Setspan=autoto automatically select span parameter (currently only recommended fortigmint-long)cut=500: Cut length for long reads (tigmint-longonly)longmap=ont: Long read platform;ontfor Oxford Nanopore Technologies (ONT) long reads,pbfor PacBio long reads (tigmint-longonly)window=1000: Window size (bp) for checking spanning moleculesminsize=2000: Minimum molecule sizeas=0.65: Minimum AS/read length rationm=5: Maximum number of mismatchesdist=50000: Maximum distance (bp) between reads to be considered the same molecule. Setdist=autoto automatically calculate dist threshold based on read length distribution (tigmint-longonly)mapq=0: Mapping quality thresholdtrim=0: Number of bases to trim off contigs following cutst=8: Number of threadsac=3000: Minimum contig length (bp) for tallying attempted corrections. This is for logging purposes only, and will not affect the performance.SORT_OPTS: specify any options to be used forsort
Parameters of ARCS
c=5e=30000r=0.05
Parameters of LINKS
a=0.1l=10
Parameters for calculating assembly metrics
ref: Reference genome,ref.fa, for calculating assembly contiguity metricsG: Size of the reference genome, for calculating NG50 and NGA50
Tips
- If your barcoded reads are in multiple FASTQ files, the initial alignments of the barcoded reads to the draft assembly can be done in parallel and merged prior to running Tigmint.
- When aligning linked reads with BWA-MEM, use the
-Coption to include the barcode in the BX tag of the alignments. - Sort by BX tag using
samtools sort -tBX. - Merge multiple BAM files using
samtools merge -tBX. - When aligning long reads with Minimap2, use the
-yoption to include the barcode in the BX tag of the alignments. - When using long reads, the minimum spanning molecule thresholds (
span) should be no greater than 1/4 of the sequence coverage. Setting the parameterspan=autoallows the appropriate parameter value to be selected automatically (this setting requires the parameterGas well). - When using long reads, the edit distance threshold (
nm) is automatically set to the cut length (cut) to compensate for the higher error rate and length. This parameter should be kept relatively high to include as many alignments as possible. - Each Tigmint (and Tigmint-long) step can be run separately through specifying the target using
tigmint-make. For example, the bwa index step for Tigmint with linked reads can be launched withtigmint-make tigmint-index- Tigmint steps\targets (for linked reads):
tigmint-index,tigmint-align,tigmint-molecule,tigmint-cut - Tigmint-long steps\targets:
tigmint-long-estimate,tigmint-long-to-linked,tigmint-long-cut
- Tigmint steps\targets (for linked reads):
- In the command line,
SORT_OPTScan be specified to add options used forsort
Using stLFR linked reads
To use stLFR linked reads with Tigmint, you will need to re-format the reads to have the barcode in a BX:Z: tag in the read header.
For example, this format
@V100002302L1C001R017000000#0_0_0/1 0 1
TGTCTTCCTGGACAGCTGACATCCCTTTTGTTTTTCTGTTTGCTCAGATGCTGTCTCTTATACACATCTTAGGAAGACAAGCACTGACGACATGATCACC
+
FFFFFFFGFGFFGFDFGFFFFFFFFFFFGFFF@FFFFFFFFFFFF@FFFFFFFFFGGFFEFEFFFF?FFFFGFFFGFFFFFFFGFFEFGFGGFGFFFGFF
should be changed to:
@V100002302L1C001R017000000 BX:Z:0_0_0
TGTCTTCCTGGACAGCTGACATCCCTTTTGTTTTTCTGTTTGCTCAGATGCTGTCTCTTATACACATCTTAGGAAGACAAGCACTGACGACATGATCACC
+
FFFFFFFGFGFFGFDFGFFFFFFFFFFFGFFF@FFFFFFFFFFFF@FFFFFFFFFGGFFEFEFFFF?FFFFGFFFGFFFFFFFGFFEFGFGGFGFFFGFF
Support
After first looking for an existing issue at https://github.com/bcgsc/tigmint/issues, please report a new issue at https://github.com/bcgsc/tigmint/issues/new. Please report the names of your input files, the exact command line that you are using, and the entire output of Tigmint.